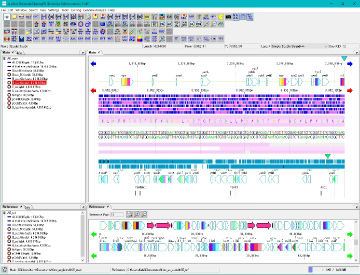
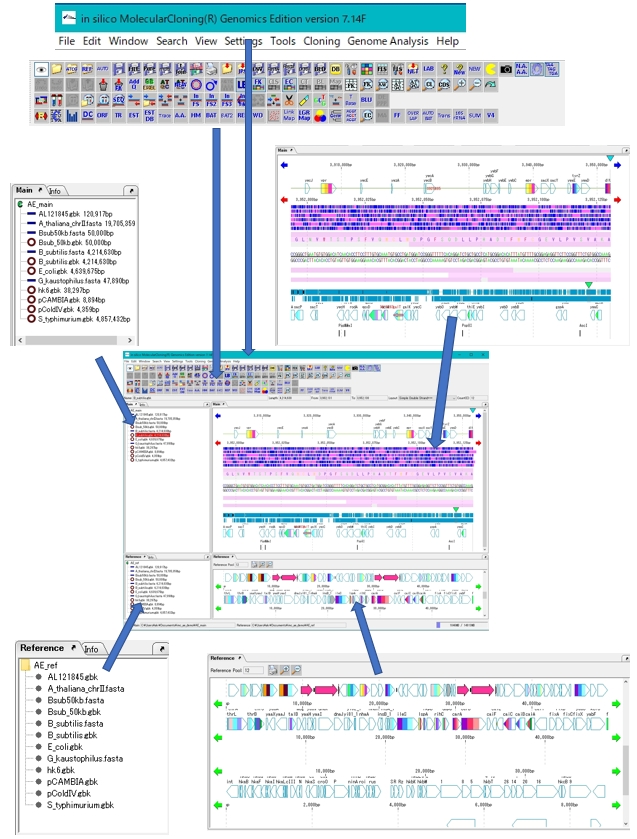
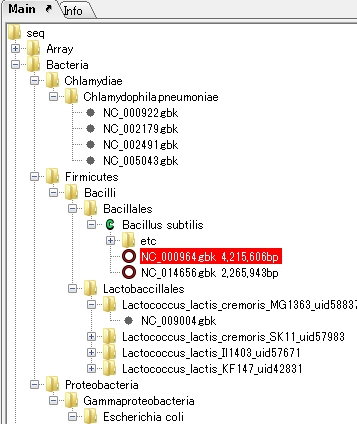
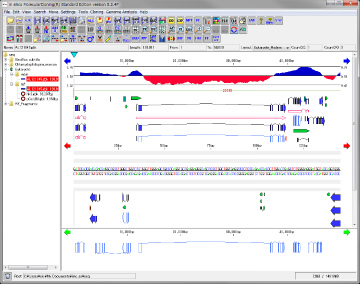
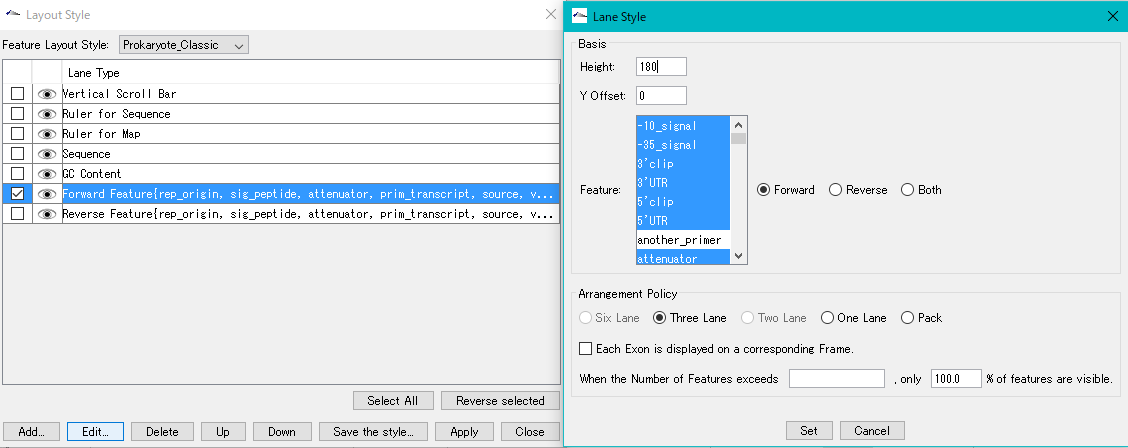
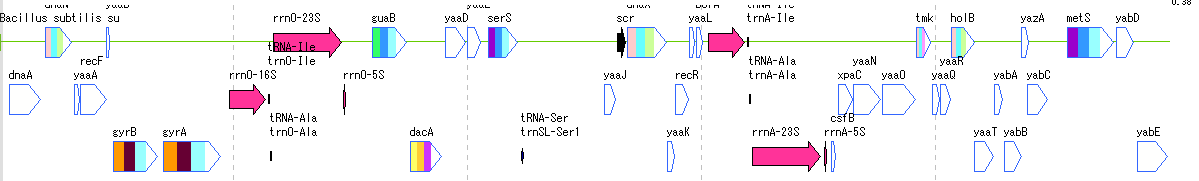
Counts the number of feature keys registered in the main current array and displays a list.
Several features to be searched can be selected, and the search range can be limited.
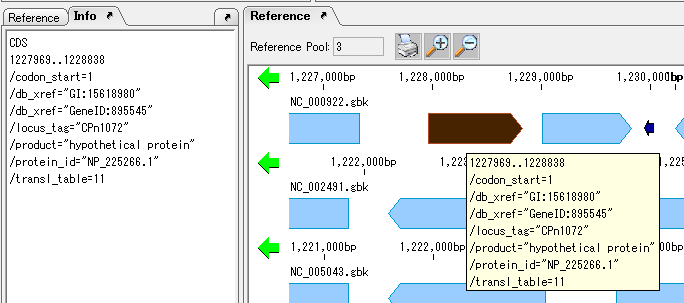
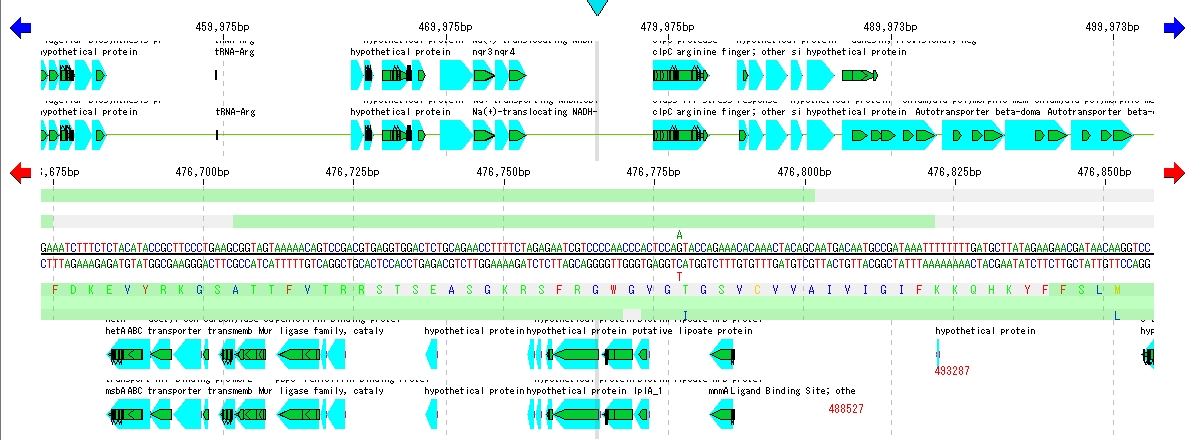
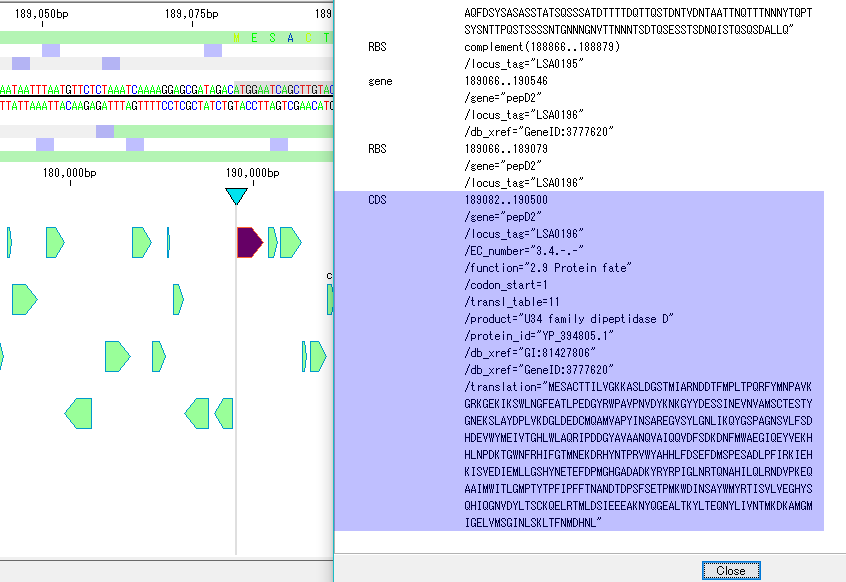
On the search result screen, buttons for feature key number, position on array, base length, gene name, base sequence, button to annotation to descriptions are listed.
In CDS, it is possible to perform operations such as selection of amino acid sequence, + chain strand only, collective deletion, codon table, serial number addition, Fusion PCR.
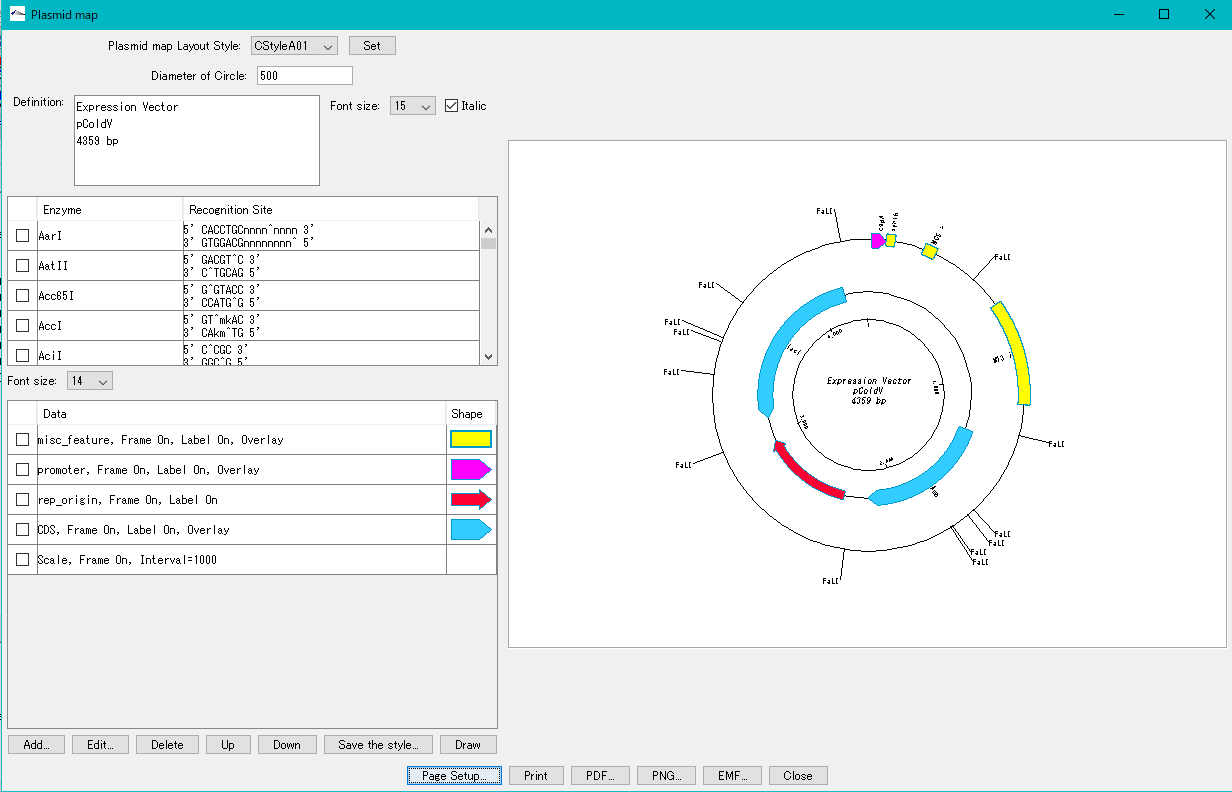
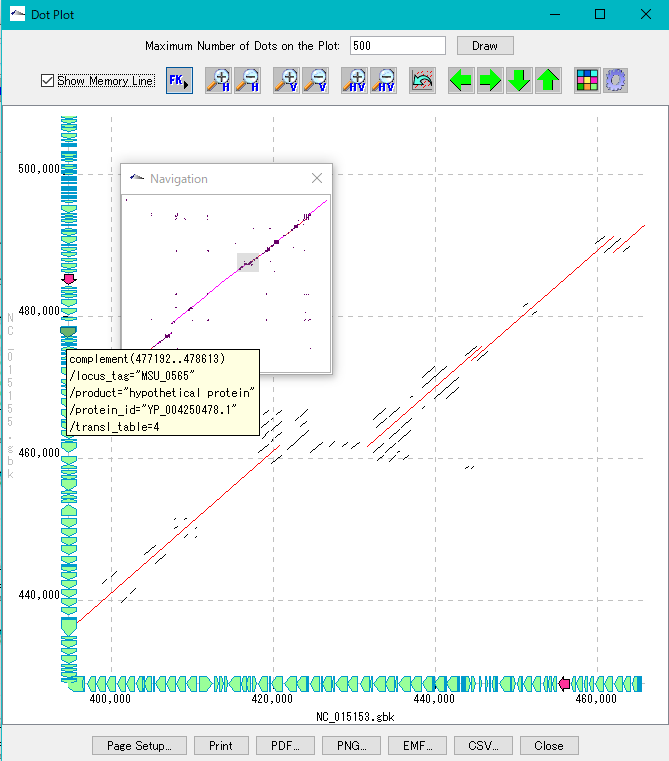
When you click a line, the main feature map shows the location of that feature.
File output in CSV / FastA / GenBank format is possible.